After my last post about visualization of 3D simulated tumors @jc_atlantis made an excellent point that another reason why 3D agent-based simulations are not performed frequently is the large amount of generated data. Of course, one approach to overcome that problem would be to store only information that is really essential and not the 1-1 simulation snapshot. However, in some cases we can’t know in advance what information will be important and thus, we need better ways to save our simulation outputs.
In this post I will show few tricks (in C++) how to efficiently store output of 2D agent-based model presented in one of the previous posts. As a result we will reduce the size of generated simulation output from about 1Gb to reasonable 35Mb. Of course, presented ideas can be utilized in 3D agent-based model and in other programing languages like Python, Java or MATLAB.
First of all let us define the task: we want to store information about spatial distribution of 1,000,000 cells on 2D lattice. Each cell is described by two variables: remaining proliferation potential (p, unsigned char variable) and if it is cancer stem cell (is_stem, boolean variable). All of the cells to be saved are stored in the STL vector. The above assumptions are defined by the following piece of the C++ code:
struct cell {//defining a cell
unsigned int place; //varible keeping linear index to cell's place on
unsigned char p; //remaining proliferation potential
bool is_stem; //is the cell a cancer stem cell?
cell(unsigned int a=0, unsigned char b=0, bool c=0):place(a), p(b), is_stem(c){}; //constructor
};
static const int N = 2000; //lattice size
vector<cell> cells; //vector containing all cells present in the system
Each cell is occupying (usigned int = 4 bytes) + (unsigned char = 1 byte) + (bool = 1 byte) = 6 bytes of memory. Hence, as we want to save 1 mln cells, our output file shouldn’t exceed 6 megabytes (Mb) of memory.
1. Outputting to text file
Information about cells is typically written to a text file in which each line describes one cell and variables values are separated by special character (standard .csv format). That format allows simulation output to be easily loaded to other programs for further analysis. Moreover, the code generating and loading the output files is very simple:
void save_cells_ASCII(string fileName) {
ofstream file_op(fileName);
for(int i=0; i<cells.size();i++)
file_op << cells.at(i).place << " " << (int)cells.at(i).p << " " << cells.at(i).is_stem << endl; file_op.close(); } void load_cells_ASCII(string fileName) { ifstream myfile (fileName); string line; unsigned int place; int p; bool is_stem; while ( getline (myfile,line) ) { istringstream iss(line); iss >> place >> p >> is_stem;
cells.push_back(cell(place, (unsigned char)p, is_stem));
}
myfile.close();
}
Code above generated a file of 11Mb of size – way more to be expected from the cell definition. Why is it so? Explanation is quite straightforward. In the generated text file each character occupies 1 byte of disk space – it is OK for the proliferation capacity (p) and boolean variable (is_stem), as they occupy the same amount of space in the memory. However, the cell position 2234521, which when stored as unsigned int occupies 4 bytes of space in the memory, occupies 7 bytes of space on the disk. In addition, each space between the written value in the text file occupies additional byte of space. All of the above together generates the wasteful 11Mb file on the disk.
2. Binary files
In order to to have a file on disk occupying exactly the same amount of space as the variables in the memory, we need to use binary files. It is a little bit more complicated to operate on binary files, but the idea is simple: we just write each variable byte by byte (1 byte = 8 bits). First, we write functions that 1) prepare array of pointers to char (byte) variables that will be saved; 2) read simulation snapshot from array of pointers to char variables.
void save_cells_binary(char** data, unsigned long* sizeData) {
unsigned long Ncells = cells.size(); //number of cells
int sizeOfCell = sizeof(int)+sizeof(char)+sizeof(bool);
*sizeData = Ncells*sizeOfCell;
*data = (char*)malloc( *sizeData );
memcpy( *data, (char*)&Ncells, sizeof(unsigned long));
memcpy((char*)&Ncells, *data, sizeof(unsigned long));
for(int i=0; i<Ncells; i++) {
memcpy(*data+i*sizeOfCell+sizeof(unsigned long), (char*)&cells.at(i).place, sizeof(int));
memcpy(*data+i*sizeOfCell+sizeof(int)+sizeof(unsigned long), (char*)&cells.at(i).p, sizeof(char));
memcpy(*data+i*sizeOfCell+sizeof(int)+sizeof(char)+sizeof(unsigned long), (char*)&cells.at(i).is_stem, sizeof(bool));
}
}
void load_cells_binary(char* data) {
unsigned long Ncells;
memcpy((char*)&Ncells, data, sizeof(unsigned long));
int sizeOfCell = sizeof(int)+sizeof(char)+sizeof(bool);
unsigned int place;
unsigned char p;
bool is_stem;
for(int i=0; i<Ncells; i++) { memcpy((char*)&place, data+sizeof(unsigned long) + i*sizeOfCell, sizeof(unsigned long)); memcpy((char*)&p, data+sizeof(unsigned long) + i*sizeOfCell+sizeof(int), sizeof(char)); memcpy((char*)&is_stem, data+sizeof(unsigned long) + i*sizeOfCell+sizeof(int)+sizeof(char), sizeof(bool)); cells.push_back(cell(place, p, is_stem)); } }
Now we need only functions that will operate on the binary files and 1) read the char array from the binary file; 2) save char array to binary file.
void readWholeBinary(string fileName, char** data, unsigned long* sizeData) { ifstream file_op(fileName, ios::binary | ios::ate); *sizeData = file_op.tellg(); file_op.seekg(0, ios::beg); *data = (char*)malloc( *sizeData ); file_op.read(*data, *sizeData); file_op.close(); } void saveWholeBinary(string fileName, char* data, unsigned long sizeData, unsigned long originalSize) { ofstream file_op(fileName, ios::binary | ios::out); if (originalSize>sizeData) //there was compression, we ass original file size to the beginning of the file
file_op.write((char*)&originalSize, sizeof(unsigned long));
file_op.write(data, sizeData);
file_op.close();
}
Executing the above code generates 5.51 Mb file on the disk (we had about 950,000 cells to save). This is about half of the space that is occupied by the text file!
3. Using zip compression
All of us probably used zip compression to store or send files through e-mail. Why won’t we write files generated by our simulation already compressed with zip? There are quite a few C++ zip libraries, but I’ve chosen zlib library as it is quite easy to use (an excellent tutorial can be found here). It operates on the char arrays that we already generate on the way to save binary files, so compressing the file before writing takes only few lines of the code.
void compressFunction (char** dataCompressed, char* dataOriginal, unsigned long* sizeDataCompressed, unsigned long sizeDataOriginal) {
*sizeDataCompressed = ((sizeDataOriginal) * 1.1) + 12;
*dataCompressed = (char*)malloc(*sizeDataCompressed);
compress((Bytef*)(*dataCompressed),sizeDataCompressed,(Bytef*)dataOriginal,sizeDataOriginal );// size of source data in bytes
}
void uncompressFunction (char** dataUncompressed, char* dataCompressed, unsigned long sizeDataCompressed, unsigned long* sizeDataUncompressed) {
memcpy((char*)sizeDataUncompressed, dataCompressed, sizeof(unsigned long));
*dataUncompressed = (char*)malloc( *sizeDataUncompressed );
uncompress((Bytef*)(*dataUncompressed), sizeDataUncompressed, (Bytef*)(dataCompressed+sizeof(unsigned long)), sizeDataCompressed-sizeof(unsigned long));
}
The binary file after compression takes 3.57 Mb of space on disk – way better.
4. Collapsing the variables
We can easily save a byte of memory per cell if we notice that the proliferation capacity variable in our simulations has the value smaller than 50. Unsigned char, however, can store the value up to 255. Thus, we can add the value 100 to the remaining proliferation capacity if the cell is cancer stem cell and forget about saving boolean variable.
memcpy(*data+i*sizeOfCell+sizeof(unsigned long), (char*)&cells.at(i).place, sizeof(int));
val = cells.at(i).p+(unsigned char)cells.at(i).is_stem*100;
memcpy(*data+i*sizeOfCell+sizeof(int)+sizeof(unsigned long), (char*)&val, sizeof(char));
5. Using information about the space to reduce the file size
The most amount of disk space is used by the information about the cells location on the lattice (unsigned int = 4 bytes). Can we skip writing that information for most of the cells? Yes, we can. In the code below we just write the cells row by row, and store the information about the location of the first cell in the lattice row. If there is an empty space between the cells in the row we put the value 255 and it the remainder of the row is empty we put value 254.
void save_cells_usingSpace(char** data, unsigned long* sizeData) {
unsigned long Ncells = cells.size(); //number of cells
int sizeOfCell = sizeof(int)+sizeof(char)+sizeof(bool);
*sizeData = 0; //we will update that value
*data = (char*)malloc( Ncells*sizeOfCell ); //we alloc more than we need
unsigned char lat[N][N] = {0};
int x, y;
for (int i = 0; i<cells.size(); i++) {
x = cells.at(i).place % N;
y = floor((double)cells.at(i).place/(double)N);
lat[x][y] = (unsigned char)cells.at(i).is_stem*(unsigned char)100 + (unsigned char)cells.at(i).p + (unsigned char)1;
}
memcpy(*data, (char*)&N, sizeof(int));
*sizeData += sizeof(int);
//255 means empty space, 254 means end of line
unsigned char es = 255, el = 254;
for (unsigned short int i = 0; i<N; i++) {
int sep = 0;
bool st = false;
for (unsigned short int j = 0; j<N; j++) { if(st == false && lat[i][j]>0) {
st = true;
memcpy(*data+*sizeData,(char*)&i, sizeof(unsigned short int));
memcpy(*data+*sizeData+sizeof(unsigned short int),(char*)&j, sizeof(unsigned short int));
memcpy(*data+*sizeData+2*sizeof(unsigned short int),(char*)&lat[i][j], sizeof(unsigned char));
*sizeData += 2*sizeof(unsigned short int)+sizeof(unsigned char);
} else if (st == true && lat[i][j] == 0 ) { //we have empty space
sep++;
} else if (st==true && lat[i][j]>0){
for (int k = 0; k<sep; k++) {
memcpy(*data+*sizeData,(char*)&es, sizeof(unsigned char));
*sizeData += sizeof(unsigned char);
}
sep = 0;
memcpy(*data+*sizeData,(char*)&lat[i][j], sizeof(unsigned char));
*sizeData += sizeof(unsigned char);
}
}
if (st == true) {
memcpy(*data+*sizeData,(char*)&el, sizeof(unsigned char));
*sizeData += sizeof(unsigned char);
}
}
}
void load_cells_usingSpace(char* data, unsigned long dataSize) {
unsigned short int x, y;
unsigned char read;
int Nm=0;
memcpy((char*)&Nm, data, sizeof(int));
unsigned long i = sizeof(int);
while(i < dataSize) {
memcpy((char*)&x,data+i, sizeof(unsigned short int));
memcpy((char*)&y,data+i+sizeof(unsigned short int), sizeof(unsigned short int));
i += 2*sizeof(unsigned short int);
unsigned int add = 0;
while (true) {
memcpy((char*)&read,data+i, sizeof(unsigned char));
i+=sizeof(unsigned char);
if (read == 254) { //end of line
break;
} else if (read < 255){//actual cell cells.push_back(cell((unsigned int)x + (unsigned int)y*Nm + add*Nm,(read-1) % 100,(read - 1)>=100));
}
add++;
}
}
}
The above approach when combined with zip compression generated file that takes only 0.47 Mb of disk space. That is less then a byte per cell!
6. Overall comparison
Fig 1. shows the amount of used space by a file written using different approaches presented above. As it can be seen the last approach uses about 25x less disk space than the standard text files approach. When saving simulation snapshots every 10 days the total amount of generated data was reduced from 1 Gb when using standard text file approach to about 35 Mb!
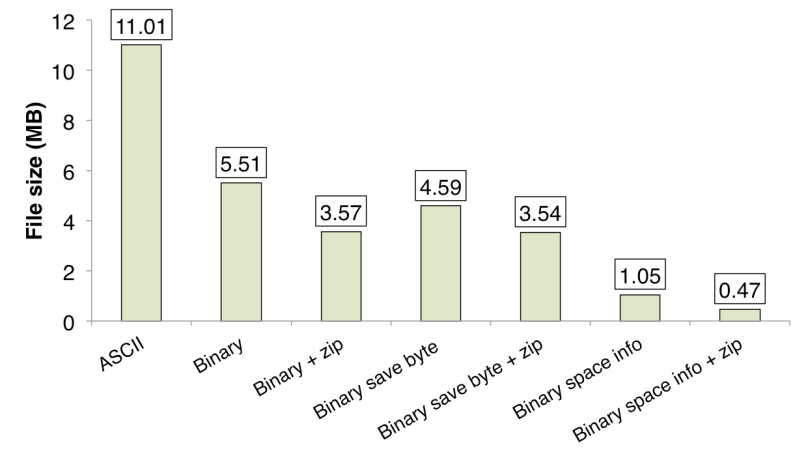 Figure 1. Comparison of the amount of used disk space.
Figure 1. Comparison of the amount of used disk space.
What is important binary files approach is way faster that text file approach, even when using zip compression algorithm, see Fig. 2.

Figure 2. File read/write speed for different save/read functions.
Of course, the trade-off is that it won’t be that easy to read our simulation files to other programs. However, we can always write wrapper functions that will allow other programs to interpret our files (see e.g. this post).


hdf5, netcdf etc ?
LikeLike
I think that it would be overkill in this case…
LikeLike