In the last couple of posts I’ve shown how to implement agent-based model of cancer stem cell driven tumor growth, both in MATLAB and C++. Having the implementations we can go one step further and perform some analysis of the tumor growth characteristics predicted by the model. We will start with performing local sensitivity analysis, i.e. we will try to answer the question how the perturbation of parameter values impacts the growth dynamics. Typically it is being done by perturbing one of the parameters by a fixed amount and analyzing the response of the model to that change. In our case the response will be the percentage change in average tumor size after 3 months of growth. Sounds fairly simple, but…
We have 5 different parameters in the model: proliferation capacity, probability of division, probability of spontaneous death, probability of symmetric division, and probability of migration. Moreover, let us assume that we would like to investigate 3 different values of parameter perturbation magnitude (5%, 10% and 20%). In order to be able to analyze the change in the average size we need to have its decent estimator, i.e. we need sufficient number of simulated stochastic trajectories – let us assume that 100 simulations is enough to have a good estimation of “true” average. So in order to perform the sensitivity analysis we will need to perform 100 + 3*5*100 = 1600 simulations (remember about the growth for nominal parameters values). Even if single simulation takes typically 30 seconds, then we will wait more than 13 hours to obtain the result using single CPU – that is a lot!
After looking at the above numbers we can make a straightforward decision right now – we will use C++ instead of MATLAB, because the model implementation in C++ is a several times faster. However 1) we will need to write a lot of a code in order to perform sensitivity analysis, 2) using multiple CPU is not that straightforward as in MATLAB. Is there a better way to proceed?
Few weeks ago I’ve shown here how to wrap your C++ in order to use it from within MATLAB as a function without loosing the C++ performance. Why not to use it to make our lives easier by making sensitivity code short and harness easily the power of multiple CPUs?
We will start the coding (in MATLAB) with setting all simulation parameters
nSim = 100; %number of simulations to perform for a given set of parameters tmax = 30*3; %number of days to simulate N = 1000; %size of the simulation domain %nominal parameter values [rhomax, pdiv, alpha, ps, pmig] nominal = [10, 1/24, 0.05, 0.3,10/24]; perturb = [0.05 0.1 0.2]; %perturbation magnitudes, percent of initial value up
Now we just need to construct a loops that will iterate through all possible perturbations and simulations. If we don’t use Parallel Toolbox, i.e. don’t use multiple CPUs, it doesn’t really matter how we will do that – performance will be similar. Otherwise, implementation is not that straightforward even though from MATLABs documentation it seems that it is enough to change for to parfor. The most important thing is how will we divide the work between the CPUs. The simples idea is to spread the considered perturbations values among the CPUs – that will allow to use 15 CPUs in our setting. However, I’ve got machine with 24 CPUs, so that would be a waste of resources – bad idea. The other idea would be to use parfor loop to spread all 100 simulations for a given perturbation value on all 24 CPUs and go through all perturbation values in a simple loop – now we are using all available CPUs. But are we doing that efficiently? No. The thing is that CPUs need to be synchronized before proceeding to the next iteration of the loop going through perturbation values. So some of the CPUs will be idle while waiting for other ones to finish with parfor loop. In order to make the code even more efficient we will just use one parfor loop and throw all 1600 simulation on 24 CPUs at the same time. Let us first prepare the output variable.
HTCG = zeros(1,nSim + length(nominal)*length(perturb)*nSim);
Before writing the final piece of the code we need to solve one more issue. Namely, in the C++ implementation we used srand(time(NULL)) to initiate the seed for random number generator. It is perfectly fine when we use single CPU – each simulation will take some time and we don’t need to worry about uniqueness of the seed. The problem is when we want to use multiple CPUs – all initial parallel simulations will start with exactly the same seed. One way to solve that is to pass the current loop iteration number (i) to C++ and use srand(time(NULL)+i) – that is what I have done. After solving that issue we can write the final piece of the code.
parfor i = 1:length(HTCG)
%%PREPARING PARAMETERS VALUES
params = nominal; %setting parameters to nominal values
if i>nSim %simulation is for perturbed parameters
%translating linear index to considered parameter and perturbation value
j = ceil((i-nSim)/(nSim*length(perturb)));
k = ceil((mod((i-nSim)-1,nSim*length(perturb))+1)/nSim);
%updating parameters
params(j) = params(j)*(1+perturb(k));
if k == 1 %if proliferation capacity parameter we need to round it
params(j) = round(params(j));
end
end
%%RUNNING SIMULATION AND SAVING OUTPUT
[~, cells] = CA(params,[tmax*24,N,i]);
HTCG(i) = length(cells)/3;
clear mex %important! without that the internal
%variables in CA functions won't be cleared and next simulation
%won't begin with single initial cell
end
Then in the command line we start the parallel pool with 24 workers (CPUs), by typing parpool(24) command, and run the code. Screenshot below shows nicely how all of the 24 CPUs are being used – no resource wasted!
We can then add few additional lines of the code to plot the results.
nom = mean(HTCG(1:100)); %average for nominal parameters value
%calculating averages for perturbed sets values
av = squeeze(mean(reshape(HTCG(101:end),nSim,length(perturb),length(nominal))));
%plotting results of sensitivity analysis
set(0,'DefaultAxesFontSize',18)
figure(1)
clf
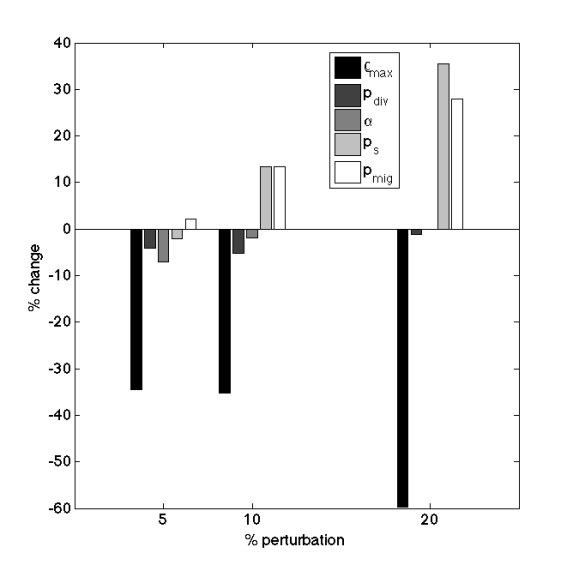
bar(perturb*100,(av-nom)/nom*100)
legend({'\rho_{max}', 'p_{div}', '\alpha', 'p_s', 'p_{mig}'})
xlabel('% perturbation')
ylabel('% change')
And “voila!” – the resulting figure shows that the perturbation in the proliferation capacity has the highest impact on the tumor growth dynamics.